Graph Neural Networks (GNNs) excel in processing data with complex relationships represented as graphs, enabling advanced analysis in social networks, molecular structures, and recommendation systems. Convolutional Neural Networks (CNNs) specialize in grid-like data such as images and videos, utilizing spatial hierarchies for effective feature extraction. Explore the differences, applications, and benefits of GNNs and CNNs to understand their unique roles in modern AI.
Why it is important
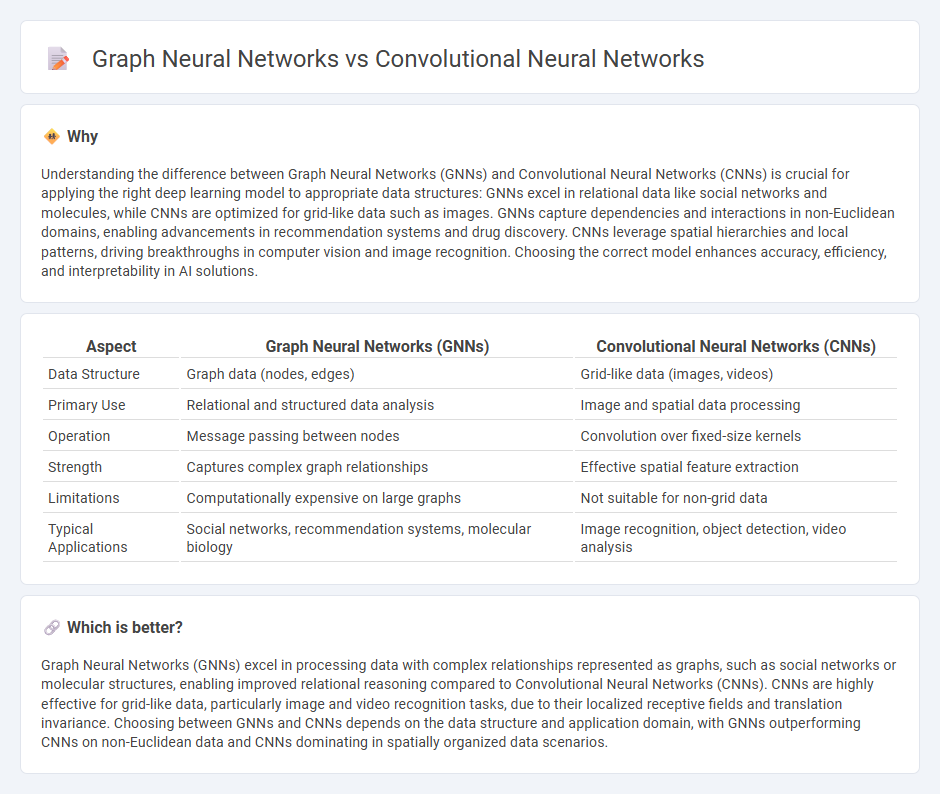
Understanding the difference between Graph Neural Networks (GNNs) and Convolutional Neural Networks (CNNs) is crucial for applying the right deep learning model to appropriate data structures: GNNs excel in relational data like social networks and molecules, while CNNs are optimized for grid-like data such as images. GNNs capture dependencies and interactions in non-Euclidean domains, enabling advancements in recommendation systems and drug discovery. CNNs leverage spatial hierarchies and local patterns, driving breakthroughs in computer vision and image recognition. Choosing the correct model enhances accuracy, efficiency, and interpretability in AI solutions.
Comparison Table
| Aspect | Graph Neural Networks (GNNs) | Convolutional Neural Networks (CNNs) |
|---|---|---|
| Data Structure | Graph data (nodes, edges) | Grid-like data (images, videos) |
| Primary Use | Relational and structured data analysis | Image and spatial data processing |
| Operation | Message passing between nodes | Convolution over fixed-size kernels |
| Strength | Captures complex graph relationships | Effective spatial feature extraction |
| Limitations | Computationally expensive on large graphs | Not suitable for non-grid data |
| Typical Applications | Social networks, recommendation systems, molecular biology | Image recognition, object detection, video analysis |
Which is better?
Graph Neural Networks (GNNs) excel in processing data with complex relationships represented as graphs, such as social networks or molecular structures, enabling improved relational reasoning compared to Convolutional Neural Networks (CNNs). CNNs are highly effective for grid-like data, particularly image and video recognition tasks, due to their localized receptive fields and translation invariance. Choosing between GNNs and CNNs depends on the data structure and application domain, with GNNs outperforming CNNs on non-Euclidean data and CNNs dominating in spatially organized data scenarios.
Connection
Graph Neural Networks (GNNs) and Convolutional Neural Networks (CNNs) share a fundamental connection through their use of local neighborhood aggregation and weight sharing mechanisms. CNNs perform convolutions on grid-structured data like images by applying filters over spatially localized pixel regions, while GNNs generalize this concept by applying convolutional operations over nodes and edges in irregular graph-structured data. This connection enables GNNs to extend the powerful feature extraction capabilities of CNNs to complex relational data in various domains such as social networks, molecular biology, and recommendation systems.
Key Terms
Feature Extraction
Convolutional Neural Networks (CNNs) excel in feature extraction from grid-like data such as images by leveraging spatial hierarchies and local connectivity patterns, enabling efficient learning of visual features. Graph Neural Networks (GNNs) specialize in extracting features from graph-structured data by aggregating information from node neighborhoods to capture complex relational dependencies. Explore how CNNs and GNNs differ in feature extraction techniques to optimize performance across various data types.
Spatial Relationships
Convolutional Neural Networks (CNNs) excel at capturing spatial hierarchies in grid-like data, making them ideal for image recognition tasks, whereas Graph Neural Networks (GNNs) specialize in learning from irregular graph-structured data with complex spatial relationships. CNNs use fixed kernel operations over local neighborhoods while GNNs aggregate information from nodes and their neighbors based on graph connectivity, enabling better modeling of non-Euclidean spaces. Explore more about how these neural network architectures handle spatial relationships for applications across computer vision and graph analytics.
Node Connectivity
Convolutional Neural Networks (CNNs) excel in processing grid-like structures such as images, leveraging local receptive fields and shared weights to capture spatial hierarchies, but they inherently assume fixed connectivity patterns. Graph Neural Networks (GNNs) specialize in modeling complex node connectivity by dynamically aggregating information from arbitrary graph structures, enabling effective representation learning in domains like social networks and molecular chemistry. Explore further to understand how node connectivity impacts different neural network architectures and their applications.
Source and External Links
An Introduction to Convolutional Neural Networks (CNNs) - Convolutional Neural Networks (CNNs) are deep learning algorithms specialized in object recognition tasks, capable of automatically extracting features from data with translation-invariance, used widely in image classification, detection, segmentation, and adaptable to domains like NLP and speech recognition through pre-trained models and fine-tuning.
Convolutional neural network - CNNs are feedforward neural networks that learn features via convolutional filters sliding over input data to produce translation-equivariant feature maps, widely applied in image/video recognition, medical image analysis, NLP, and financial time series, inspired by the biological visual cortex's receptive fields and requiring less preprocessing than traditional methods.
Convolutional Neural Networks (CNNs / ConvNets) - CNNs are neural networks with learnable weights and biases structured specifically for images; they differ from ordinary networks by encoding image properties into their architecture, making them efficient with fewer parameters while mapping raw pixels to class scores through convolutional and fully connected layers.
 dowidth.com
dowidth.com